API¶
Limbo follows a policy-based design, which allows users to combine high flexibility (almost every part of Limbo can be substituted by a user-defined part) with high performance (the abstraction do not add any overhead, contrary to classic OOP design). These two features are critical for researchers who want to experiment new ideas in Bayesian optimization. This means that changing a part of limbo (e.g. changing the kernel functions) usually corresponds to changing a template parameter of the optimizer.
The parameters of the algorithms (e.g. an epsilon) are given by a template class (usually called Params in our code, and always the first argument). See Parameters for details.
To avoid defining each component of an optimizer manually, Limbo provides sensible defaults. In addition, Limbo relies on Boost.Parameter to make it easy to customize a single part. This Boost library allows us to write classes that accept template argument (user-defined custom classes) by name. For instance, to customize the stopping criteria:
using namespace limbo;
// here stop_t is a user-defined list of stopping criteria
bayes_opt::BOptimizer<Params, stopcrit<stop_t>> boptimizer;
Or to define a custom acquisition function:
using namespace limbo;
// here acqui_t is a user-defined acquisition function
bayes_opt::BOptimizer<Params, acquifun<acqui_t>> boptimizer;
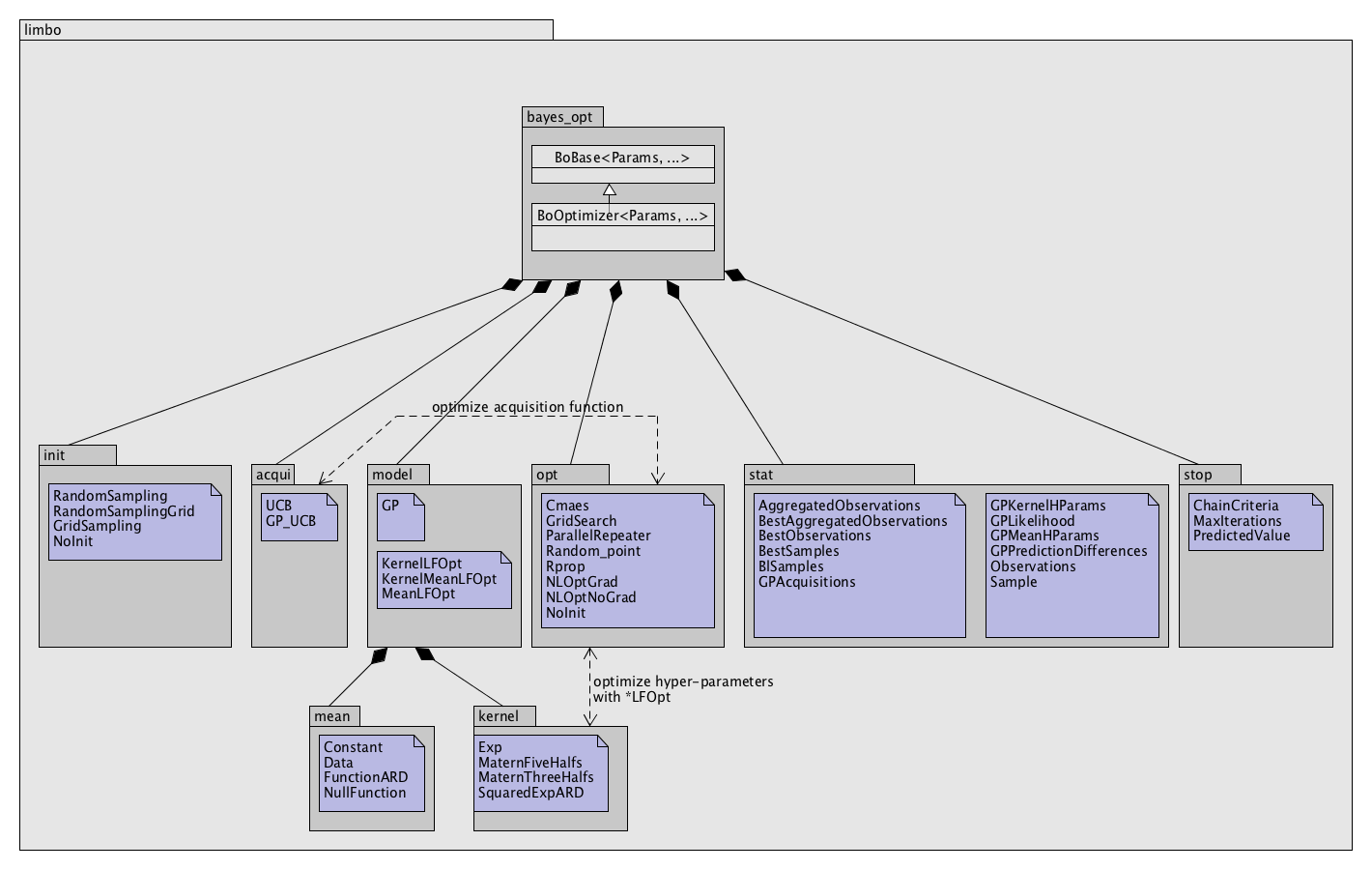
Class Structure¶
There is almost no explicit inheritance in Limbo because polymorphism is not used. However, each kind of class follow a similar template (or ‘concept’), that is, they have to implement the same methods. For instance, every initialization function must implement a operator() method:
template <typename StateFunction, typename AggregatorFunction, typename Opt>
void operator()(const StateFunction& seval, const AggregatorFunction&, Opt& opt) const
However, there is no need to inherit from a particular ‘abstract’ class.
Every class is parametrized by a Params class that contains all the parameters.
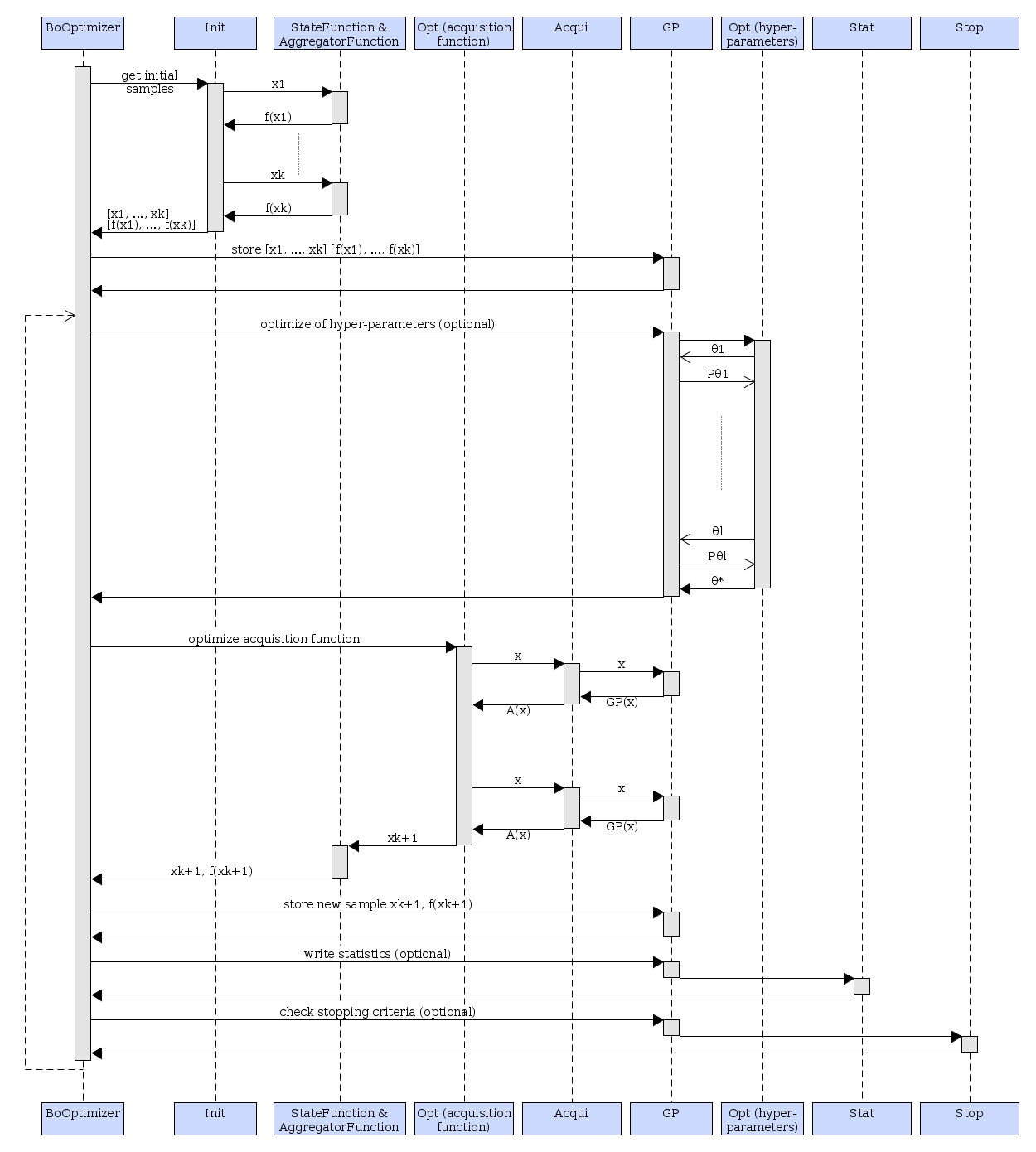
Sequence diagram¶
File Structure¶
(see below for a short explanation of the concepts)
src
+-- limbo:
|-- acqui: acquisition functions
|-- bayes_opt: bayesian optimizers
|-- init: initialization functions
|-- kernel: kernel functions
|-- mean: mean functions
|-- model: models (Gaussian Processes)
|-- opt: optimizers (Rprop, CMA-ES, etc.)
|-- stat: statistics (to dump data)
|-- stop: stopping criteria
|-- tools: useful macros & small functions
|-- tests: unit tests
|-- benchmarks: a few benchmark functions
|-- examples: a few examples
|-- cmaes: [external] the CMA-ES library,
used for inner optimizations -- from https://www.lri.fr/~hansen/cmaesintro.html
|-- ehvi: [external] the Expected HyperVolume Improvement,
used for Multi-Objective Optimization -- by Iris Hupkens
Each directory in the limbo directory corresponds to a namespace with the same name. There is also a file for each directory called “directory.hpp” (e.g. acqui.hpp) that includes the whole namespace.
Bayesian optimizers (bayes_opt)¶
- template <class Params, class A1 = boost::parameter::void_, class A2 = boost::parameter::void_, class A3 = boost::parameter::void_, class A4 = boost::parameter::void_, class A5 = boost::parameter::void_, class A6 = boost::parameter::void_>
-
class
limbo::bayes_opt::BoBase¶ Base class for Bayesian optimizers
- Parameters:
bool Params::bayes_opt_bobase::stats_enabled: activate / deactivate the statistics
This class is templated by several types with default values (thanks to boost::parameters).
type typedef argument default init. func. init_t initfun RandomSampling model model_t modelfun GP<…> acquisition fun. aqui_t acquifun GP_UCB statistics stat_t statfun see below stopping crit. stop_t stopcrit MaxIterations For GP, the default value is:
model::GP<Params, kf_t, mean_t, opt_t>>,- with
kf_t = kernel::SquaredExpARD<Params> - with
mean_t = mean::Data<Params> - with
opt_t = model::gp::KernelLFOpt<Params>
(meaning: kernel with automatic relevance determination and mean equals to the mean of the input data, that is, center the data automatically)
For Statistics, the default value is:
boost::fusion::vector<stat::Samples<Params>, stat::AggregatedObservations<Params>, stat::ConsoleSummary<Params>>Example of customization:
using Kernel_t = kernel::MaternFiveHalves<Params>;using Mean_t = mean::Data<Params>;using GP_t = model::GP<Params, Kernel_t, Mean_t>;using Acqui_t = acqui::UCB<Params, GP_t>;bayes_opt::BOptimizer<Params, modelfun<GP_t>, acquifun<Acqui_t>> opt;
Subclassed by limbo::bayes_opt::BOptimizer< Params, A1, A2, A3, A4, A5, A6 >, limbo::experimental::bayes_opt::BoMulti< Params, A1, A2, A3, A4, A5, A6 >, limbo::experimental::bayes_opt::CBOptimizer< Params, A1, A2, A3, A4, A5, A6, A7 >
Public Functions
-
BoBase()¶ default constructor
-
bool
stats_enabled() const¶ return true if the statitics are enabled (they can be disabled to avoid dumping data, e.g. for unit tests)
-
const std::string &
res_dir() const¶ return the name of the directory in which results (statistics) are written
-
const std::vector<Eigen::VectorXd> &
observations() const¶ return the vector of points of observations (observations can be multi-dimensional, hence the VectorXd) f(x)
-
const std::vector<Eigen::VectorXd> &
samples() const¶ return the list of the points that have been evaluated so far (x)
-
int
current_iteration() const¶ return the current iteration number
-
void
add_new_sample(const Eigen::VectorXd &s, const Eigen::VectorXd &v)¶ Add a new sample / observation pair
- does not update the model!
- we don’t add NaN and inf observations
- template <typename StateFunction>
-
void
eval_and_add(const StateFunction &seval, const Eigen::VectorXd &sample)¶ Evaluate a sample and add the result to the ‘database’ (sample / observations vectors) it does not update the model.
- template <class Params, class A1 = boost::parameter::void_, class A2 = boost::parameter::void_, class A3 = boost::parameter::void_, class A4 = boost::parameter::void_, class A5 = boost::parameter::void_, class A6 = boost::parameter::void_>
-
class
limbo::bayes_opt::BOptimizer¶ The classic Bayesian optimization algorithm.
References: [brochu2010tutorial][Mockus2013]
This class takes the same template parameters as BoBase. It adds:
type typedef argument default acqui. optimizer acquiopt_t acquiopt see below The default value of acqui_opt_t is:
opt::NLOptNoGrad<Params, nlopt::GN_DIRECT_L_RAND>if NLOpt was found inwaf configureopt::Cmaes<Params>if libcmaes was found but NLOpt was not foundopt::GridSearch<Params>otherwise (please do not use this: the algorithm will not work as expected!)
Inherits from limbo::bayes_opt::BoBase< Params, A1, A2, A3, A4, A5, A6 >
Public Types
Public Functions
- template <typename StateFunction, typename AggregatorFunction = FirstElem>
-
void
optimize(const StateFunction &sfun, const AggregatorFunction &afun = AggregatorFunction(), bool reset = true)¶ The main function (run the Bayesian optimization algorithm)
- template <typename AggregatorFunction = FirstElem>
-
const Eigen::VectorXd &
best_observation(const AggregatorFunction &afun = AggregatorFunction()) const¶ return the best observation so far (i.e. max(f(x)))
- template <typename AggregatorFunction = FirstElem>
-
const Eigen::VectorXd &
best_sample(const AggregatorFunction &afun = AggregatorFunction()) const¶ return the best sample so far (i.e. the argmax(f(x)))
Acquisition functions (acqui)¶
An acquisition function is what is optimized to select the next point to try. It usually depends on the model.
Template¶
template <typename Params, typename Model>
class AcquiName {
public:
AcquiName(const Model& model, int iteration = 0) : _model(model) {}
size_t dim_in() const { return _model.dim_in(); }
size_t dim_out() const { return _model.dim_out(); }
template <typename AggregatorFunction>
limbo::opt::eval_t operator()(const Eigen::VectorXd& v, const AggregatorFunction& afun, bool gradient) const
{
// code
}
};
Available acquisition functions¶
-
group
acqui - template <typename Params, typename Model>
-
class
EI¶ - #include <limbo/acqui/ei.hpp>
Classic EI (Expected Improvement). See [brochu2010tutorial], p. 14
\[\begin{split}EI(x) = (\mu(x) - f(x^+) - \xi)\Phi(Z) + \sigma(x)\phi(Z),\\\text{with } Z = \frac{\mu(x)-f(x^+) - \xi}{\sigma(x)}.\end{split}\]- Parameters:
double jitter- \(\xi\)
- template <typename Params, typename Model>
-
class
GP_UCB¶ - #include <limbo/acqui/gp_ucb.hpp>
GP-UCB (Upper Confidence Bound). See [brochu2010tutorial], p. 15. See also: http://arxiv.org/abs/0912.3995
\[UCB(x) = \mu(x) + \kappa \sigma(x).\]with:
\[\kappa = \sqrt{2 \log{(\frac{n^{D/2+2}\pi^2}{3 \delta})}}\]where \(n\) is the number of past evaluations of the objective function and \(D\) the dimensionality of the parameters (dim_in).
- Parameters:
- double delta (a small number in [0,1], e.g. 0.1)
- template <typename Params, typename Model>
-
class
UCB¶ - #include <limbo/acqui/ucb.hpp>
Classic UCB (Upper Confidence Bound). See [brochu2010tutorial], p. 14
\[UCB(x) = \mu(x) + \alpha \sigma(x).\]- Parameters:
double alpha
Default Parameters¶
-
group
Acqui_defaults Functions
-
limbo::defaults::acqui_ei::BO_PARAM(double, jitter, 0. 0)
-
limbo::defaults::acqui_gpucb::BO_PARAM(double, delta, 0. 1)
-
limbo::defaults::acqui_ucb::BO_PARAM(double, alpha, 0. 5)
-
limbo::defaults::acqui_eci::BO_PARAM(double, jitter, 0. 0)
-
Init functions (init)¶
Initialization functions are used to inialize a Bayesian optimization algorithm with a few samples. For instance, we typically start with a dozen of random samples.
Template¶
struct InitName {
template <typename StateFunction, typename AggregatorFunction, typename Opt>
void operator()(const StateFunction& seval, const AggregatorFunction&, Opt& opt) const
{
// code
}
Available initializers¶
-
group
init - template <typename Params>
-
struct
GridSampling¶ - #include <limbo/init/grid_sampling.hpp>
Grid sampling.
- Parameter:
int bins(number of bins)
- template <typename Params>
-
struct
LHS¶ - #include <limbo/init/lhs.hpp>
Latin Hypercube sampling in [0, 1]^n (LHS)
- Parameters:
int samples(total number of samples)
- template <typename Params>
-
struct
NoInit¶ - #include <limbo/init/no_init.hpp>
Do nothing (dummy initializer).
- template <typename Params>
-
struct
RandomSampling¶ - #include <limbo/init/random_sampling.hpp>
Pure random sampling in [0, 1]^n
- Parameters:
int samples(total number of samples)
- template <typename Params>
-
struct
RandomSamplingGrid¶ - #include <limbo/init/random_sampling_grid.hpp>
Grid-based random sampling: in effect, define a grid and takes random samples from this grid. The number of bins in the grid is ``bins“^number_of_dimensions
For instance, if bins = 5 and there are 3 inputs dimensions, then the grid is 5*5*5=125, and random points will all be points of this grid.
- Parameters:
int samples(total number of samples)int bins(number of bins for each dimensions)
Default Parameters¶
-
group
init_defaults Functions
-
limbo::defaults::init_gridsampling::BO_PARAM(int, bins, 5)
-
limbo::defaults::init_lhs::BO_PARAM(int, samples, 10)
-
limbo::defaults::init_randomsampling::BO_PARAM(int, samples, 10)
-
limbo::defaults::init_randomsamplinggrid::BO_PARAM(int, samples, 10)
-
limbo::defaults::init_randomsamplinggrid::BO_PARAM(int, bins, 5)
-
Optimization functions (opt)¶
In Limbo, optimizers are used both to optimize acquisition functions and to optimize hyper-parameters. However, this API might be helpful in other places whenever an optimization of a function is needed.
Warning
Limbo optimizers always MAXIMIZE f(x), whereas many libraries MINIMIZE f(x)
Most algorithms are wrappers to external libraries (NLOpt and CMA-ES). Only the Rprop (and a few control algorithms like ‘RandomPoint’) is implemented in Limbo. Some optimizers require the gradient, some don’t.
The tutorial Optimization sub-API describes how to use the opt:: API in your own algorithms.
The return type of the function to be optimized is eval_t, which is defined as a pair of a double (f(x)) and a vector (the gradient):
using eval_t = std::pair<double, boost::optional<Eigen::VectorXd>>;
To make it easy to work with eval_t, Limbo defines a few shortcuts:
-
group
opt_tools Typedefs
-
using
limbo::opt::eval_t = typedef std::pair<double, boost::optional<Eigen::VectorXd>> return type of the function to optimize
Functions
-
eval_t
no_grad(double x)¶ return with opt::no_grad(your_val) if no gradient is available (to be used in functions to be optimized)
-
const Eigen::VectorXd &
grad(const eval_t &fg)¶ get the gradient from a function evaluation (eval_t)
-
double
fun(const eval_t &fg)¶ get the value from a function evaluation (eval_t)
- template <typename F>
-
double
eval(const F &f, const Eigen::VectorXd &x)¶ Evaluate f without gradient (to be called from the optimization algorithms that do not use the gradient)
- template <typename F>
-
eval_t
eval_grad(const F &f, const Eigen::VectorXd &x)¶ Evaluate f with gradient (to be called from the optimization algorithms that use the gradient)
-
using
Template¶
template <typename Params>
struct OptimizerName {
template <typename F>
Eigen::VectorXd operator()(const F& f, const Eigen::VectorXd& init, bool bounded) const
{
// content
}
};
fis the function to be optimized. If the gradient is known, the function should look like this:
limbo::opt::eval_t my_function(const Eigen::VectorXd& v, bool eval_grad = false)
{
double fx = <function_value>;
Eigen::VectorXd gradient = <gradient>;
return {fx, gradient};
}
It is possible to make it a bit more generic by not computing the gradient when it is not asked, that is:
limbo::opt::eval_t my_function(const Eigen::VectorXd& v, bool eval_grad = false)
{
double fx = <function_value>;
if (!eval_grad)
return opt::no_grad(v);
Eigen::VectorXd gradient = <gradient>;
return {fx, gradient};
}
- If the gradient of
fis not known:
limbo::opt::eval_t my_function(const Eigen::VectorXd& v, bool eval_grad = false)
{
double x = <function_value>(v);
return limbo::opt::no_grad(x);
}
initis an optionnal starting point (for local optimizers); many optimizers ignore this argument (see the table below): in that case, an assert will fail.boundedis true if the optimization is bounded in [0,1]; many optimizers do not support bounded optimization (see the table below).eval_gradallows Limbo to avoid computing the gradient when it is not needed (i.e. when the gradient is known but we optimize using a gradient-free optimizer).
To call an optimizer (e.g. NLOptGrad):
// the type of the optimizer (here NLOpt with the LN_LBGFGS algorithm)
opt::NLOptGrad<ParamsGrad, nlopt::LD_LBFGS> lbfgs;
// we start from a random point (in 2D), and the search is not bounded
Eigen::VectorXd res_lbfgs = lbfgs(my_function, tools::random_vector(2), false);
std::cout <<"Result with LBFGS:\t" << res_lbfgs.transpose()
<< " -> " << my_function(res_lbfgs).first << std::endl;
Not all the algorithms support bounded optimization and/or initial point:
| Algo. | bounded | init |
|---|---|---|
| CMA-ES | yes | yes |
| NLOptGrad | * | * |
| NLOptNoGrad | * | * |
| Rprop | no | yes |
| RandomPoint | yes | no |
* All NLOpt’s global optimizers must have bounds. Check NLOpt’s reference to see which algorithms support initial point.
Available optimizers¶
-
group
opt - template <typename Params>
-
struct
Cmaes¶ - #include <limbo/opt/cmaes.hpp>
Covariance Matrix Adaptation Evolution Strategy by Hansen et al. (See: https://www.lri.fr/~hansen/cmaesintro.html)
- our implementation is based on libcmaes (https://github.com/beniz/libcmaes)
- Support bounded and unbounded optimization
- Only available if libcmaes is installed (see the compilation instructions)
- Parameters :
- int variant
- int elitism
- int restarts
- double max_fun_evals
- double fun_tolerance
- double xrel_tolerance
- double fun_target
- bool fun_compute_initial
- bool handle_uncertainty
- bool verbose
- double lb (lower bounds)
- double ub (upper bounds)
- template <typename Params>
-
struct
GridSearch¶ - #include <limbo/opt/grid_search.hpp>
Grid search
Parameters:
- int bins
- template <typename Params, nlopt::algorithm Algorithm = nlopt::LD_LBFGS>
-
struct
NLOptGrad¶ - #include <limbo/opt/nlopt_grad.hpp>
Binding to gradient-based NLOpt algorithms. See: http://ab-initio.mit.edu/wiki/index.php/NLopt_Algorithms
Algorithms:
- GD_STOGO
- GD_STOGO_RAND
- LD_LBFGS_NOCEDAL
- LD_LBFGS
- LD_VAR1
- LD_VAR2
- LD_TNEWTON
- LD_TNEWTON_RESTART
- LD_TNEWTON_PRECOND
- LD_TNEWTON_PRECOND_RESTART
- GD_MLSL
- GD_MLSL_LDS
- LD_MMA
- LD_AUGLAG
- LD_AUGLAG_EQ
- LD_SLSQP
- LD_CCSAQ
Parameters :
- int iterations
- double fun_tolerance
- double xrel_tolerance
- template <typename Params, nlopt::algorithm Algorithm = nlopt::GN_DIRECT_L_RAND>
-
struct
NLOptNoGrad¶ - #include <limbo/opt/nlopt_no_grad.hpp>
Binding to gradient-free NLOpt algorithms. See: http://ab-initio.mit.edu/wiki/index.php/NLopt_Algorithms
Algorithms:
- GN_DIRECT
- GN_DIRECT_L, [default]
- GN_DIRECT_L_RAND
- GN_DIRECT_NOSCAL
- GN_DIRECT_L_NOSCAL
- GN_DIRECT_L_RAND_NOSCAL
- GN_ORIG_DIRECT
- GN_ORIG_DIRECT_L
- GN_CRS2_LM
- GN_MLSL
- GN_MLSL_LDS
- GN_ISRES
- LN_COBYLA
- LN_AUGLAG_EQ
- LN_BOBYQA
- LN_NEWUOA
- LN_NEWUOA_BOUND
- LN_PRAXIS
- LN_NELDERMEAD
- LN_SBPLX
- LN_AUGLAG
Parameters:
- int iterations
- double fun_tolerance
- double xrel_tolerance
- template <typename Params, typename Optimizer>
-
struct
ParallelRepeater¶ - #include <limbo/opt/parallel_repeater.hpp>
Meta-optimizer: run the same algorithm in parallel many times from different init points and return the maximum found among all the replicates (useful for local algorithms)
Parameters:
- int repeats
- template <typename Params>
-
struct
RandomPoint¶ - #include <limbo/opt/random_point.hpp>
- return a random point in [0, 1]
- no parameters
- useful for control experiments (do not use this otherwise!)
- template <typename Params>
-
struct
Rprop¶ - #include <limbo/opt/rprop.hpp>
Gradient-based optimization (rprop)
- partly inspired by libgp: https://github.com/mblum/libgp
- reference : Blum, M., & Riedmiller, M. (2013). Optimization of Gaussian Process Hyperparameters using Rprop. In European Symposium on Artificial Neural Networks, Computational Intelligence and Machine Learning.
Parameters:
- int iterations
Default parameters¶
-
group
opt_defaults Functions
-
limbo::defaults::opt_cmaes::BO_PARAM(int, restarts, 1) number of restarts of CMA-ES
-
limbo::defaults::opt_cmaes::BO_PARAM(double, max_fun_evals, - 1) maximum number of calls to the function to be optimized
-
limbo::defaults::opt_cmaes::BO_PARAM(double, fun_tolerance, - 1) threshold based on the difference in value of a fixed number of trials: if bigger than 0, it enables the tolerance criteria for stopping based in the history of rewards.
-
limbo::defaults::opt_cmaes::BO_PARAM(double, xrel_tolerance, - 1) tolerance for convergence: stop when an optimization step (or an estimate of the optimum) changes all the parameter values by less than tol multiplied by the absolute value of the parameter value. IGNORED if negative
-
limbo::defaults::opt_cmaes::BO_PARAM(double, fun_target, - 1) function value target: if bigger than 0, enables the function target criteria for stopping if the performance is greater than this value.
-
limbo::defaults::opt_cmaes::BO_PARAM(bool, fun_compute_initial, false) computes initial objective function value: if true, it evaluates the provided starting point (if any).
-
BO_PARAM(int, variant, aIPOP_CMAES)¶ sets the version of cmaes to use (possible values are: CMAES_DEFAULT, IPOP_CMAES, BIPOP_CMAES, aCMAES, aIPOP_CMAES, aBIPOP_CMAES, sepCMAES, sepIPOP_CMAES, sepBIPOP_CMAES, sepaCMAES, sepaIPOP_CMAES, sepaBIPOP_CMAES, VD_CMAES, VD_IPOP_CMAES, VD_BIPOP_CMAES
-
limbo::defaults::opt_cmaes::BO_PARAM(int, elitism, 0) defines elitism strategy: 0 -> no elitism 1 -> elitism: reinjects the best-ever seen solution 2 -> initial elitism: reinject x0 as long as it is not improved upon 3 -> initial elitism on restart: restart if best encountered solution is not the the final solution and reinjects the best solution until the population has better fitness, in its majority
-
limbo::defaults::opt_cmaes::BO_PARAM(bool, handle_uncertainty, false) enables or disables uncertainty handling: https://hal.inria.fr/file/index/docid/276216/filename/TEC2008.pdf
-
limbo::defaults::opt_cmaes::BO_PARAM(bool, verbose, false) enables or disables verbose mode for cmaes
-
limbo::defaults::opt_cmaes::BO_PARAM(double, lbound, 0. 0) lower bound (in input) for cmaes
-
limbo::defaults::opt_cmaes::BO_PARAM(double, ubound, 1. 0) upper bound (in input) for cmaes
-
limbo::defaults::opt_gridsearch::BO_PARAM(int, bins, 5) number of bins for each dimension
-
limbo::defaults::opt_nloptgrad::BO_PARAM(int, iterations, 500) number of calls to the optimized function
-
limbo::defaults::opt_nloptgrad::BO_PARAM(double, fun_tolerance, - 1) tolerance for convergence: stop when an optimization step (or an estimate of the optimum) changes the objective function value by less than tol multiplied by the absolute value of the function value. IGNORED if negative
-
limbo::defaults::opt_nloptgrad::BO_PARAM(double, xrel_tolerance, - 1) tolerance for convergence: stop when an optimization step (or an estimate of the optimum) changes all the parameter values by less than tol multiplied by the absolute value of the parameter value. IGNORED if negative
-
limbo::defaults::opt_nloptnograd::BO_PARAM(int, iterations, 500) number of calls to the optimized function
-
limbo::defaults::opt_nloptnograd::BO_PARAM(double, fun_tolerance, - 1) tolerance for convergence: stop when an optimization step (or an estimate of the optimum) changes the objective function value by less than tol multiplied by the absolute value of the function value. IGNORED if negative
-
limbo::defaults::opt_nloptnograd::BO_PARAM(double, xrel_tolerance, - 1) tolerance for convergence: stop when an optimization step (or an estimate of the optimum) changes all the parameter values by less than tol multiplied by the absolute value of the parameter value. IGNORED if negative
-
limbo::defaults::opt_parallelrepeater::BO_PARAM(int, repeats, 10) number of replicates
-
limbo::defaults::opt_rprop::BO_PARAM(int, iterations, 300) number of max iterations
-
Models / Gaussian processes (model)¶
Currently, Limbo only includes Gaussian processes as models. More may come in the future.
- template <typename Params, typename KernelFunction = kernel::MaternFiveHalves<Params>, typename MeanFunction = mean::Data<Params>, typename HyperParamsOptimizer = gp::NoLFOpt<Params>>
-
class
limbo::model::GP¶ A classic Gaussian process. It is parametrized by:
- a kernel function
- a mean function
- [optionnal] an optimizer for the hyper-parameters
Public Functions
-
GP()¶ useful because the model might be created before knowing anything about the process
-
GP(int dim_in, int dim_out)¶ useful because the model might be created before having samples
-
void
compute(const std::vector<Eigen::VectorXd> &samples, const std::vector<Eigen::VectorXd> &observations, bool compute_kernel = true)¶ Compute the GP from samples and observations. This call needs to be explicit!
-
void
optimize_hyperparams()¶ Do not forget to call this if you use hyper-prameters optimization!!
-
void
add_sample(const Eigen::VectorXd &sample, const Eigen::VectorXd &observation)¶ add sample and update the GP. This code uses an incremental implementation of the Cholesky decomposition. It is therefore much faster than a call to compute()
-
std::tuple<Eigen::VectorXd, double>
query(const Eigen::VectorXd &v) const¶ \rst return :math:
\mu, :math:\sigma^2(unormalized). If there is no sample, return the value according to the mean function. Using this method instead of separate calls to mu() and sigma() is more efficient because some computations are shared between mu() and sigma(). \endrst
-
Eigen::VectorXd
mu(const Eigen::VectorXd &v) const¶ \rst return :math:
\mu(unormalized). If there is no sample, return the value according to the mean function. \endrst
-
double
sigma(const Eigen::VectorXd &v) const¶ \rst return :math:
\sigma^2(unormalized). If there is no sample, return the max :math:\sigma^2. \endrst
-
int
dim_in() const¶ return the number of dimensions of the input
-
int
dim_out() const¶ return the number of dimensions of the output
-
Eigen::VectorXd
max_observation() const¶ return the maximum observation (only call this if the output of the GP is of dimension 1)
-
Eigen::VectorXd
mean_observation() const¶ return the mean observation (only call this if the output of the GP is of dimension 1)
-
double
compute_log_lik()¶ compute and return the log likelihood
-
Eigen::VectorXd
compute_kernel_grad_log_lik()¶ compute and return the gradient of the log likelihood wrt to the kernel parameters
-
Eigen::VectorXd
compute_mean_grad_log_lik()¶ compute and return the gradient of the log likelihood wrt to the mean parameters
-
double
get_log_lik() const¶ return the likelihood (do not compute it return last computed)
-
void
set_log_lik(double log_lik)¶ set the log likelihood (e.g. computed from outside)
-
double
compute_log_loo_cv()¶ compute and return the log probability of LOO CV
-
Eigen::VectorXd
compute_kernel_grad_log_loo_cv()¶ compute and return the gradient of the log probability of LOO CV wrt to the kernel parameters
-
double
get_log_loo_cv() const¶ return the LOO-CV log probability (do not compute it return last computed)
-
void
set_log_loo_cv(double log_loo_cv)¶ set the LOO-CV log probability (e.g. computed from outside)
-
const Eigen::MatrixXd &
matrixL() const¶ LLT matrix (from Cholesky decomposition)
-
const std::vector<Eigen::VectorXd> &
samples() const¶ return the list of samples that have been tested so far
- template <typename A>
-
void
save(const std::string &directory)¶ save the parameters and the data for the GP to the archive (text or binary)
- template <typename A>
-
void
save(const A &archive)¶ save the parameters and the data for the GP to the archive (text or binary)
The hyper-parameters of the model (kernel, mean) can be optimized. The following options are possible:
-
group
model_opt - template <typename Params, typename Optimizer = opt::ParallelRepeater<Params, opt::Rprop<Params>>>
-
struct
HPOpt¶ - #include <limbo/model/gp/hp_opt.hpp>
optimize the likelihood of the kernel only
Subclassed by limbo::model::gp::KernelLFOpt< Params, Optimizer >, limbo::model::gp::KernelLooOpt< Params, Optimizer >, limbo::model::gp::KernelMeanLFOpt< Params, Optimizer >, limbo::model::gp::MeanLFOpt< Params, Optimizer >
- template <typename Params, typename Optimizer = opt::ParallelRepeater<Params, opt::Rprop<Params>>>
-
struct
KernelLFOpt¶ - #include <limbo/model/gp/kernel_lf_opt.hpp>
optimize the likelihood of the kernel only
Inherits from limbo::model::gp::HPOpt< Params, Optimizer >
- template <typename Params, typename Optimizer = opt::ParallelRepeater<Params, opt::Rprop<Params>>>
-
struct
KernelLooOpt¶ - #include <limbo/model/gp/kernel_loo_opt.hpp>
optimize the likelihood of the kernel only
Inherits from limbo::model::gp::HPOpt< Params, Optimizer >
- template <typename Params, typename Optimizer = opt::ParallelRepeater<Params, opt::Rprop<Params>>>
-
struct
KernelMeanLFOpt¶ - #include <limbo/model/gp/kernel_mean_lf_opt.hpp>
optimize the likelihood of both the kernel and the mean (try to align the mean function)
Inherits from limbo::model::gp::HPOpt< Params, Optimizer >
- template <typename Params, typename Optimizer = opt::ParallelRepeater<Params, opt::Rprop<Params>>>
-
struct
MeanLFOpt¶ - #include <limbo/model/gp/mean_lf_opt.hpp>
optimize the likelihood of the mean only (try to align the mean function)
Inherits from limbo::model::gp::HPOpt< Params, Optimizer >
- template <typename Params>
-
struct
NoLFOpt¶ - #include <limbo/model/gp/no_lf_opt.hpp>
do not optimize anything
See the Gaussian Process tutorial for a tutorial about using GP without using a Bayesian optimization algorithm.
Kernel functions (kernel)¶
Template¶
template <typename Params>
struct Kernel : public BaseKernel<Params, Kernel<Params>> {
Kernel(size_t dim = 1) {}
double kernel(const Eigen::VectorXd& v1, const Eigen::VectorXd& v2) const
{
// code
}
};
Available kernels¶
-
group
kernel - template <typename Params>
-
struct
Exp¶ - #include <limbo/kernel/exp.hpp>
Exponential kernel (see [brochu2010tutorial] p. 9).
\[k(v_1, v_2) = \sigma^2\exp \Big(-\frac{||v_1 - v_2||^2}{2l^2}\Big)\]- Parameters:
double sigma_sq(signal variance)double l(characteristic length scale)
Inherits from limbo::kernel::BaseKernel< Params, Exp< Params > >
- template <typename Params, typename Kernel>
-
struct
BaseKernel¶ - #include <limbo/kernel/kernel.hpp>
Base struct for kernel definition. It handles the noise and its optimization (only if the kernel allows hyper-parameters optimization).
Parameters:
double noise(initial signal noise squared)bool optimize_noise(whether we are optimizing for the noise or not)
- template <typename Params>
-
struct
MaternFiveHalves¶ - #include <limbo/kernel/matern_five_halves.hpp>
Matern kernel
\[ \begin{align}\begin{aligned}d = ||v1 - v2||\\\nu = 5/2\\C(d) = \sigma^2\frac{2^{1-\nu}}{\Gamma(\nu)}\Bigg(\sqrt{2\nu}\frac{d}{l}\Bigg)^\nu K_\nu\Bigg(\sqrt{2\nu}\frac{d}{l}\Bigg),\end{aligned}\end{align} \]- Parameters:
double sigma_sq(signal variance)double l(characteristic length scale)
Reference: [matern1960spatial] & [brochu2010tutorial] p.10 & https://en.wikipedia.org/wiki/Mat%C3%A9rn_covariance_function
Inherits from limbo::kernel::BaseKernel< Params, MaternFiveHalves< Params > >
- template <typename Params>
-
struct
MaternThreeHalves¶ - #include <limbo/kernel/matern_three_halves.hpp>
Matern 3/2 kernel
\[ \begin{align}\begin{aligned}d = ||v1 - v2||\\\nu = 3/2\\C(d) = \sigma^2\frac{2^{1-\nu}}{\Gamma(\nu)}\Bigg(\sqrt{2\nu}\frac{d}{l}\Bigg)^\nu K_\nu\Bigg(\sqrt{2\nu}\frac{d}{l}\Bigg),\end{aligned}\end{align} \]- Parameters:
double sigma_sq(signal variance)double l(characteristic length scale)
Reference: [matern1960spatial] & [brochu2010tutorial] p.10 & https://en.wikipedia.org/wiki/Mat%C3%A9rn_covariance_function
Inherits from limbo::kernel::BaseKernel< Params, MaternThreeHalves< Params > >
- template <typename Params>
-
struct
SquaredExpARD¶ - #include <limbo/kernel/squared_exp_ard.hpp>
Squared exponential covariance function with automatic relevance detection (to be used with a likelihood optimizer) Computes the squared exponential covariance like this:
\[k_{SE}(v1, v2) = \sigma^2 \exp \Big(-\frac{1}{2}(v1-v2)^TM(v1-v2)\Big),\]with \(M = \Lambda\Lambda^T + diag(l_1^{-2}, \dots, l_n^{-2})\) being the characteristic length scales and \(\alpha\) describing the variability of the latent function. The parameters \(l_1^2, \dots, l_n^2, \Lambda,\sigma^2\) are expected in this order in the parameter array. \(\Lambda\) is a \(D\times k\) matrix with \(k<D\).
- Parameters:
double sigma_sq(initial signal variance)int k(number of columns of \(\Lambda\) matrix)
Reference: [Rasmussen2006], p. 106 & [brochu2010tutorial], p. 10
Inherits from limbo::kernel::BaseKernel< Params, SquaredExpARD< Params > >
Default parameters¶
-
group
Kernel_defaults Functions
-
limbo::defaults::kernel_exp::BO_PARAM(double, sigma_sq, 1)
-
limbo::defaults::kernel::BO_PARAM(double, noise, 0. 01)
-
limbo::defaults::kernel_maternfivehalves::BO_PARAM(double, sigma_sq, 1)
-
limbo::defaults::kernel_maternfivehalves::BO_PARAM(double, l, 1)
-
limbo::defaults::kernel_maternthreehalves::BO_PARAM(double, sigma_sq, 1)
-
limbo::defaults::kernel_maternthreehalves::BO_PARAM(double, l, 1)
-
limbo::defaults::kernel_squared_exp_ard::BO_PARAM(int, k, 0)
-
limbo::defaults::kernel_squared_exp_ard::BO_PARAM(double, sigma_sq, 1)
-
Mean functions (mean)¶
Mean functions capture the prior about the function to be optimized.
Template¶
template <typename Params>
struct MeanFunction : public BaseMean<Params> {
MeanFunction(size_t dim_out = 1) : _dim_out(dim_out) {}
template <typename GP>
Eigen::VectorXd operator()(const Eigen::VectorXd& v, const GP&) const
{
// code
}
protected:
size_t _dim_out;
};
Available mean functions¶
-
group
mean - template <typename Params>
-
struct
Constant¶ - #include <limbo/mean/constant.hpp>
A constant mean (the traditionnal choice for Bayesian optimization)
Parameter:
double constant(the value of the constant)
Inherits from limbo::mean::BaseMean< Params >
- template <typename Params>
-
struct
Data¶ - #include <limbo/mean/data.hpp>
Use the mean of the observation as a constant mean
Inherits from limbo::mean::BaseMean< Params >
- template <typename Params>
-
struct
BaseMean¶ - #include <limbo/mean/mean.hpp>
Base struct for mean definition.
Subclassed by limbo::mean::Constant< Params >, limbo::mean::Data< Params >, limbo::mean::FunctionARD< Params, MeanFunction >, limbo::mean::NullFunction< Params >
- template <typename Params>
-
struct
NullFunction¶ - #include <limbo/mean/null_function.hpp>
Constant with m=0
Inherits from limbo::mean::BaseMean< Params >
Default parameters¶
-
group
mean_defaults Functions
-
limbo::defaults::mean_constant::BO_PARAM(double, constant, 1)
-
Internals¶
- template <typename Params, typename MeanFunction>
-
struct
limbo::mean::FunctionARD¶ Functor used to optimize the mean function using the maximum likelihood principle
It incorporates the hyperparameters of the underlying mean function, if any
Inherits from limbo::mean::BaseMean< Params >
Stopping criteria (stop)¶
Stopping criteria are used to stop the Bayesian optimizer algorithm.
Template¶
template <typename Params>
struct Name {
template <typename BO, typename AggregatorFunction>
bool operator()(const BO& bo, const AggregatorFunction&)
{
// return true if stop
}
};
Available stopping criteria¶
-
group
stop - template <typename Params>
-
struct
MaxIterations¶ - #include <limbo/stop/max_iterations.hpp>
Stop after a given number of iterations
parameter: int iterations
- template <typename Params, typename Optimizer = boost::parameter::void_>
-
struct
MaxPredictedValue¶ - #include <limbo/stop/max_predicted_value.hpp>
Stop once the value for the best sample is above : ratio * (best value predicted by the model)
Parameter: double ratio
Default parameters¶
-
group
stop_defaults Functions
-
limbo::defaults::stop_maxiterations::BO_PARAM(int, iterations, 190)
-
limbo::defaults::stop_maxpredictedvalue::BO_PARAM(double, ratio, 0. 9)
-
Internals¶
- template <typename BO, typename AggregatorFunction>
-
struct
limbo::stop::ChainCriteria¶ Utility functor for boost::fusion::accumulate, e.g.:
stop::ChainCriteria<BO, AggregatorFunction> chain(bo, afun); return boost::fusion::accumulate(_stopping_criteria, false, chain);
Where
_stopping_criteria` is a ``boost::fusion::vectorof classes.
Statistics (stats)¶
Statistics are used to report informations about the current state of the algorithm (e.g., the best observation for each iteration). They are typically chained in a boost::fusion::vector<>.
Template¶
template <typename Params>
struct Samples : public StatBase<Params> {
template <typename BO, typename AggregatorFunction>
void operator()(const BO& bo, const AggregatorFunction&)
{
// code
}
};
- template <typename Params>
-
struct
limbo::stat::StatBase¶ Base class for statistics
The only method provided is protected :
template <typename BO> void _create_log_file(const BO& bo, const std::string& name)
This method allocates an attribute _log_file (type: std::shared_ptr<std::ofstream>) if it has not been created yet, and does nothing otherwise. This method is designed so that you can safely call it in operator() while being ‘guaranteed’ that the file exists. Using this method is not mandatory for a statistics class.
Subclassed by limbo::experimental::stat::HyperVolume< Params >, limbo::experimental::stat::ParetoFront< Params >, limbo::stat::AggregatedObservations< Params >, limbo::stat::BestAggregatedObservations< Params >, limbo::stat::BestObservations< Params >, limbo::stat::BestSamples< Params >, limbo::stat::ConsoleSummary< Params >, limbo::stat::GP< Params >, limbo::stat::GPAcquisitions< Params >, limbo::stat::GPKernelHParams< Params >, limbo::stat::GPLikelihood< Params >, limbo::stat::GPMeanHParams< Params >, limbo::stat::GPPredictionDifferences< Params >, limbo::stat::Observations< Params >, limbo::stat::Samples< Params >
Available statistics¶
-
group
stat - template <typename Params>
-
struct
AggregatedObservations¶ - #include <limbo/stat/aggregated_observations.hpp>
TODO fallocati Write all the observations filename:
aggregated_observations.datInherits from limbo::stat::StatBase< Params >
- template <typename Params>
-
struct
BestAggregatedObservations¶ - #include <limbo/stat/best_aggregated_observations.hpp>
TODO fallocati Write the best observation at each iteration filename:
best_aggregated_observations.datInherits from limbo::stat::StatBase< Params >
- template <typename Params>
-
struct
BestObservations¶ - #include <limbo/stat/best_observations.hpp>
Write the best observation so far filename:
best_observations.dat"Inherits from limbo::stat::StatBase< Params >
- template <typename Params>
-
struct
BestSamples¶ - #include <limbo/stat/best_samples.hpp>
filename:
best_samples.datInherits from limbo::stat::StatBase< Params >
- template <typename Params>
-
struct
ConsoleSummary¶ - #include <limbo/stat/console_summary.hpp>
write the status of the algorithm on the terminal
Inherits from limbo::stat::StatBase< Params >
- template <typename Params>
-
struct
GP¶ - #include <limbo/stat/gp.hpp>
filename:
gp_<iteration>.datInherits from limbo::stat::StatBase< Params >
- template <typename Params>
-
struct
GPAcquisitions¶ - #include <limbo/stat/gp_acquisitions.hpp>
filename:
gp_acquisitions.datInherits from limbo::stat::StatBase< Params >
- template <typename Params>
-
struct
GPKernelHParams¶ - #include <limbo/stat/gp_kernel_hparams.hpp>
filename:
gp_kernel_hparams.datInherits from limbo::stat::StatBase< Params >
- template <typename Params>
-
struct
GPLikelihood¶ - #include <limbo/stat/gp_likelihood.hpp>
filename:
gp_likelihood.datInherits from limbo::stat::StatBase< Params >
- template <typename Params>
-
struct
GPMeanHParams¶ - #include <limbo/stat/gp_mean_hparams.hpp>
filename:
gp_mean_hparams.datInherits from limbo::stat::StatBase< Params >
- template <typename Params>
-
struct
GPPredictionDifferences¶ - #include <limbo/stat/gp_prediction_differences.hpp>
filename:
gp_prediction_differences.datInherits from limbo::stat::StatBase< Params >
- template <typename Params>
-
struct
Observations¶ - #include <limbo/stat/observations.hpp>
filename:
observations.datInherits from limbo::stat::StatBase< Params >
- template <typename Params>
-
struct
Samples¶ - #include <limbo/stat/samples.hpp>
filename:
samples.datInherits from limbo::stat::StatBase< Params >
Default parameters¶
-
group
stat_defaults
Parallel tools (par)¶
-
namespace
limbo::tools::par¶ Typedefs
-
using
limbo::tools::par::vector = typedef std::vector<X>
Functions
- template <typename V>
-
V
convert_vector(const V &v)¶
-
void
init()¶ init TBB (if activated) for multi-core computing
- template <typename F>
-
void
loop(size_t begin, size_t end, const F &f)¶ parallel for
- template <typename Iterator, typename F>
-
void
for_each(Iterator begin, Iterator end, const F &f)¶ parallel for_each
- template <typename T, typename F, typename C>
-
T
max(const T &init, int num_steps, const F &f, const C &comp)¶ parallel max
- template <typename T1, typename T2, typename T3>
-
void
sort(T1 i1, T2 i2, T3 comp)¶ parallel sort
- template <typename F>
-
void
replicate(size_t nb, const F &f)¶ replicate a function nb times
-
using
Misc tools (tools)¶
-
namespace
limbo::tools¶ Typedefs
-
using
limbo::tools::rdist_double_t = typedef std::uniform_real_distribution<double>
-
using
limbo::tools::rdist_int_t = typedef std::uniform_int_distribution<int>
-
using
limbo::tools::rdist_gauss_t = typedef std::normal_distribution<>
-
using
limbo::tools::rgen_double_t = typedef RandomGenerator<rdist_double_t> Double random number generator
-
using
limbo::tools::rgen_gauss_t = typedef RandomGenerator<rdist_gauss_t> Double random number generator (gaussian)
-
using
limbo::tools::rgen_int_t = typedef RandomGenerator<rdist_int_t> integer random number generator
Functions
-
Eigen::VectorXd
make_vector(double x)¶ make a 1-D vector from a double (useful when we need to return vectors)
- template <typename T>
-
constexpr int
signum(T x, std::false_type is_signed)¶
- template <typename T>
-
constexpr int
signum(T x, std::true_type is_signed)¶
- template <typename T>
-
constexpr int
signum(T x)¶ return -1 if x < 0; return 0 if x = 0; return 1 if x > 0.
- template <typename T, typename std::enable_if< std::is_arithmetic< T >::value, int >::type = 0>
-
bool
is_nan_or_inf(T v)¶ return true if v is nan (not a number) or infinity
- template <typename T, typename std::enable_if<!std::is_arithmetic< T >::value, int >::type = 0>
-
bool
is_nan_or_inf(const T &v)¶ return true if v is nan (not a number) or infinity (const version)
-
Eigen::VectorXd
random_vector_bounded(int size)¶ random vector in [0, 1.0]
- this function is thread safe because we use a random generator for each thread
- we use a C++11 random number generator
-
Eigen::VectorXd
random_vector_unbounded(int size)¶ random vector generated with a normal distribution centered on 0, with standard deviation of 10.0
- this function is thread safe because we use a random generator for each thread
- we use a C++11 random number generator
-
Eigen::VectorXd
random_vector(int size, bool bounded = true)¶ random vector wrapper for both bounded and unbounded versions
-
Eigen::MatrixXd
random_lhs(int dim, int n)¶ generate n random samples with Latin Hypercube Sampling (LHS) in [0, 1]^dim
-
std::string
date()¶ easy way to get the current date
-
std::string
hostname()¶ easy way to get the hostame
-
std::string
getpid()¶ easy way to get the PID
- template <typename D>
-
class
RandomGenerator¶ - #include <limbo/tools/random_generator.hpp>
a mt19937-based random generator (mutex-protected)
usage :
- RandomGenerator<dist<double>>(0.0, 1.0);
- double r = rgen.rand();
-
namespace
par¶ Typedefs
-
using
limbo::tools::par::vector = typedef std::vector<X>
Functions
- template <typename V>
-
V
convert_vector(const V &v)¶
-
void
init()¶ init TBB (if activated) for multi-core computing
- template <typename F>
-
void
loop(size_t begin, size_t end, const F &f)¶ parallel for
- template <typename Iterator, typename F>
-
void
for_each(Iterator begin, Iterator end, const F &f)¶ parallel for_each
- template <typename T, typename F, typename C>
-
T
max(const T &init, int num_steps, const F &f, const C &comp)¶ parallel max
- template <typename T1, typename T2, typename T3>
-
void
sort(T1 i1, T2 i2, T3 comp)¶ parallel sort
- template <typename F>
-
void
replicate(size_t nb, const F &f)¶ replicate a function nb times
-
using
-
using